GenBank Type Provider
This article describes how to use the GenBank Type Provider to remotely access genomic data stored in the GenBank database. This Type Provider collects and parses the genomic data for a specified organism and generates a static type containing its metadata and sequence.
The GenBank Type Provider uses .NET Bio to parse the GenBank data files and BioFSharp to provide utilities for manipulating genomic sequences.
Loading BioProviders Package
To load the GenBank Type Provider, a script can use the NuGet syntax to reference the BioProviders package, shown below.
You can optionally include the BioFSharp package. While it's not required to use the basic BioProviders functions, it can be used to explore the metadata of the provided types, as shown in a later example.
#r "nuget: BioProviders"
#r "nuget: BioFSharp"
If creating an F# library or application, BioProviders can be added as a package reference. You can use your IDE for this, or use the dotnet add package BioProviders command in your project folder from the command line.
BioProviders can then be used in your script or code by using an open command. Opening its dependencies should not be required. (BioFSharp is loaded for future examples.)
open BioProviders
open BioFSharp
GenBankProvider Example
The GenBank Type Provider will be demonstrated for this GenBank assembly of the Candidatus Carsonella ruddii species. To create a typed representation of the assembly, two pieces of information must be given to the Type Provider:
- Species name
- GenBank assembly accession
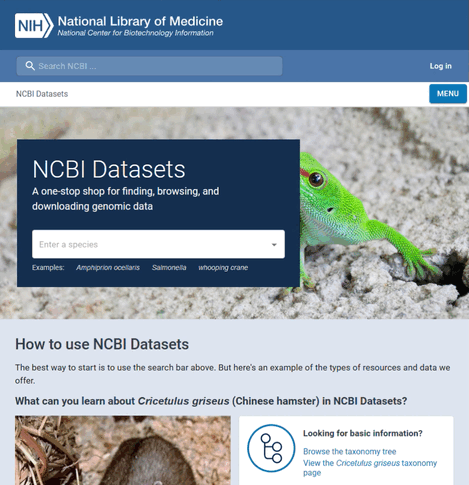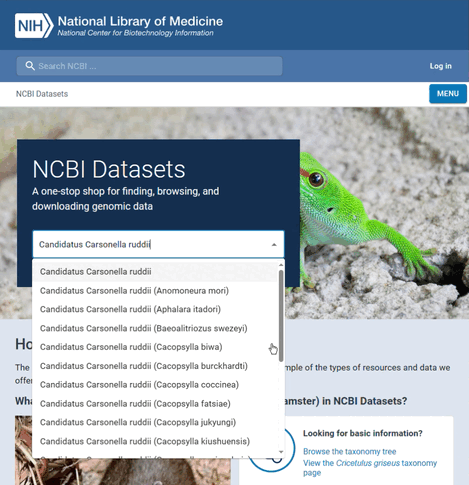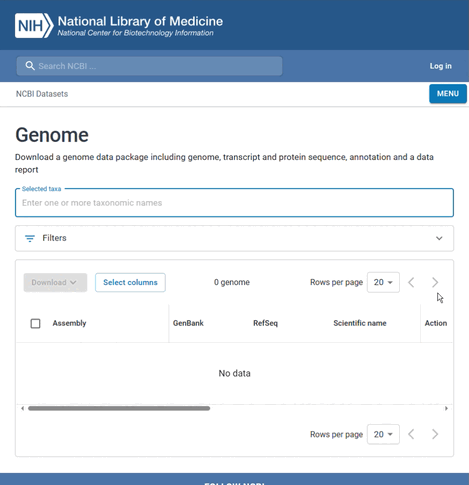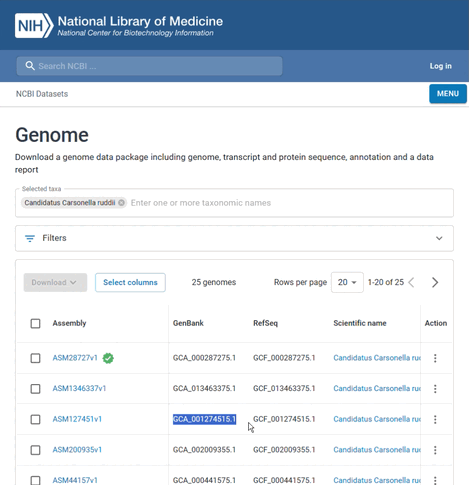
For this example, the species name is "Candidatus Carsonella ruddii" and the GenBank assembly accession is "GCA_001274515.1". To find this information:
- Visit https://www.ncbi.nlm.nih.gov/datasets/
- Search for the name of the species
- Select to view all genones of the species
You can then select the assembly's GenBank (as well as RefSeq) accession from the list that appears.
 .
.
Passing this information to the Type Provider generates the Assembly Type. The genomic data can then be extracted from the Assembly Type by invoking the Genome method. This is demonstrated below.
// Define species name and GenBank assembly accession.
let [<Literal>] Species = "Candidatus Carsonella ruddii"
let [<Literal>] Accession = "GCA_001274515.1"
// Create GenBank assembly type.
type Ruddii = GenBankProvider<Species, Accession>
// Extract statically-typed genome data.
let genome = Ruddii.Genome()
Metadata
Each genome is accompanied by metadata describing the organism and sequence recorded in the assembly. This metadata can be extracted using the Metadata field of the Genome Type created previously. The Metadata type is largely based on that provided by .NET Bio, with modifications made to be more idiomatic with F#.
Below is an example of how the raw metadata type can be retrieved and displayed:
// Extract the metadata.
let metadata = genome.Metadata
// Display the metadata type.
printf "%A" metadata
|
The metadata type consists of many fields, though not all fields of the metadata exist for all assemblies. Therefore, they are provided as option types, on which a match expression can be used. Below are examples of accessing fields from the example assembly. ✅ Example - Accessing a field that is provided.
// Print definition if exists.
match metadata.Definition with
| Some definition -> printf "%s" definition
| None -> printf "No definition provided."
|
❌ Example - Accessing a field that is not provided.
// Print database source if exists.
match metadata.DbSource with
| Some dbsource -> printf "%s" dbsource
| None -> printf "No database source provided."
|
Sequence
The genomic sequence for the organism can be extracted using the Sequence field of the Genome Type created previously. This field provides a BioFSharp BioSeq containing a series of Nucleotides. More can be read about BioFSharp containers here.
An example of accessing and manipulating the GenBankProvider genomic sequence using BioFSharp is provided below:
// Extract the BioFSharp BioSeq.
let sequence = genome.Sequence
// Display the sequence type.
printf "%A" sequence
|
// Take the complement, then transcribe and translate the coding strand.
sequence
|> BioSeq.complement
|> BioSeq.transcribeCodingStrand
|> BioSeq.translate 0
|
Wildcard Operators
Wildcard operators are supported in both the Species and Accession provided to the GenBankProvider. By using asterisks "*" at the end of a Species or Accession name, species or accessions starting with the provided pattern will be matched.
For example, we can get all Staphylococcus species starting with the letter 'c' and assembly accesions starting with 'GCA_01':
// Define species name and GenBank assembly accession using wildcards.
let [<Literal>] SpeciesPattern = "Staphylococcus c*"
let [<Literal>] AccessionPattern = "GCA_01*"
// Create GenBank type containing all species matching the species pattern.
type SpeciesCollection = GenBankProvider<SpeciesPattern, AccessionPattern>
// Select the species types.
type Capitis = SpeciesCollection.``Staphylococcus capitis``
type Cohnii = SpeciesCollection.``Staphylococcus cohnii``
// Select assemblies.
type Assembly1 = Capitis.``GCA_012926605.1``
type Assembly2 = Capitis.``GCA_015645205.1``
type Assembly3 = Cohnii.``GCA_013349225.1``
type Assembly4 = Cohnii.``GCA_014884245.1``
// Extract statically-typed genome data.
let data = Assembly1.Genome()
// Show the assembly's definition.
match data.Metadata.Definition with
| Some definition -> printf "%s" definition
| None -> printf "No definition provided."
|
The Accession parameter can also be omitted from the GenBankProvider. In this case, all assemblies for the given species will be matched. For example:
// Define species name.
let [<Literal>] SpeciesName = "Staphylococcus lugdunensis"
// Create GenBank type containing all assemblies for the species.
type Assemblies = GenBankProvider<SpeciesName>
// Select assemblies.
type Assembly = Assemblies.``GCA_001546615.1``
// Show the assembly's primary accession.
match (Assembly.Genome()).Metadata.Accession with
| Some accession -> match accession.Primary with
| Some primary -> printf "%s" primary
| None -> printf "No primary accession provided."
| None -> printf "No accession provided."
|
type LiteralAttribute = inherit Attribute new: unit -> LiteralAttribute
--------------------
new: unit -> LiteralAttribute
<summary>Typed representation of the NCBI FTP server, for GenBank data.</summary> <param name="Species">The name of the species whose genome is being accessed (e.g. "Staphylococcus borealis"). Defaults to <c>""</c>.</param> <param name="Accession">The accession of the genome assembly being accessed (e.g. "GCA_003042555.1"). Defaults to <c>""</c>.</param>
<summary>Typed representation of an Assembly's Genomic GenBank Flat File.</summary>
GenBankFlatFile.GenBankFlatFile.Metadata: Metadata.Metadata
--------------------
property GenBankProvider<...>.Genome.Metadata: Metadata.Metadata with get
<summary>Typed representation of the Metadata of a Genomic GenBank Flat File.</summary>
GenBankFlatFile.GenBankFlatFile.Sequence: BioSeq.BioSeq<Nucleotides.Nucleotide>
--------------------
property GenBankProvider<...>.Genome.Sequence: System.Collections.Generic.IEnumerable<Nucleotides.Nucleotide> with get
<summary>Typed representation of the Sequence of a Genomic GenBank Flat File.</summary>
GenBankFlatFile.GenBankFlatFile.Metadata: Metadata.Metadata
--------------------
property GenBankProvider<...>.Staphylococcus capitis.GCA_012926605.1.Genome.Metadata: Metadata.Metadata with get
<summary>Typed representation of the Metadata of a Genomic GenBank Flat File.</summary>
<summary> Identifier assigned to each GenBank sequence record. </summary>

